1 2 3 import pandas as pdimport numpy as npimport matplotlib.pyplot as plt
数据集描述 蘑菇数据集,包括毒性,大小,表面,颜色等,所有数据均为字符串类型,分析毒性与其他属性的关系
读取数据集 1 2 dataset = pd.read_csv("./mushrooms.csv" ) dataset.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 8124 entries, 0 to 8123
Data columns (total 23 columns):
class 8124 non-null object
cap-shape 8124 non-null object
cap-surface 8124 non-null object
cap-color 8124 non-null object
bruises 8124 non-null object
odor 8124 non-null object
gill-attachment 8124 non-null object
gill-spacing 8124 non-null object
gill-size 8124 non-null object
gill-color 8124 non-null object
stalk-shape 8124 non-null object
stalk-root 8124 non-null object
stalk-surface-above-ring 8124 non-null object
stalk-surface-below-ring 8124 non-null object
stalk-color-above-ring 8124 non-null object
stalk-color-below-ring 8124 non-null object
veil-type 8124 non-null object
veil-color 8124 non-null object
ring-number 8124 non-null object
ring-type 8124 non-null object
spore-print-color 8124 non-null object
population 8124 non-null object
habitat 8124 non-null object
dtypes: object(23)
memory usage: 1.4+ MB
可以发现,一共包括23个属性,没有缺失值
直观分析——颜色鲜艳的蘑菇都有毒? 1 2 3 4 5 6 7 8 poison = dataset[dataset["class" ] == "p" ]["cap-color" ] not_poison = dataset[dataset["class" ] != "p" ]["cap-color" ] poison_color = pd.concat([pd.value_counts(poison),pd.value_counts(not_poison),pd.value_counts(dataset["cap-color" ])],axis=1 ,keys=["poison" ,"normal" ,"all" ]) poison_color = poison_color.fillna(value=0 ) poison_color = poison_color.groupby(poison_color.columns,axis=1 ).apply(lambda x:x / x.sum ()) print(poison_color.sort_values(by="poison" ).loc[["p" ,"b" ,"y" ,"e" ]])
poison normal all
p 0.022472 0.013308 0.017725
b 0.030644 0.011407 0.020679
y 0.171604 0.095057 0.131955
e 0.223698 0.148289 0.184638
可得还是有一定道理的,尤其是黄色和红色的蘑菇
相关性分析——判断各指标与毒性相关性 计算各不同指标下有毒的概率判断单独指标与毒性之间的关系
1 2 3 4 5 def analysis_poison (data,index_name ): data["class" ].replace({"p" :1 ,"e" :0 },inplace=True ) return data.groupby([index_name])["class" ].sum () / pd.value_counts(data[index_name])
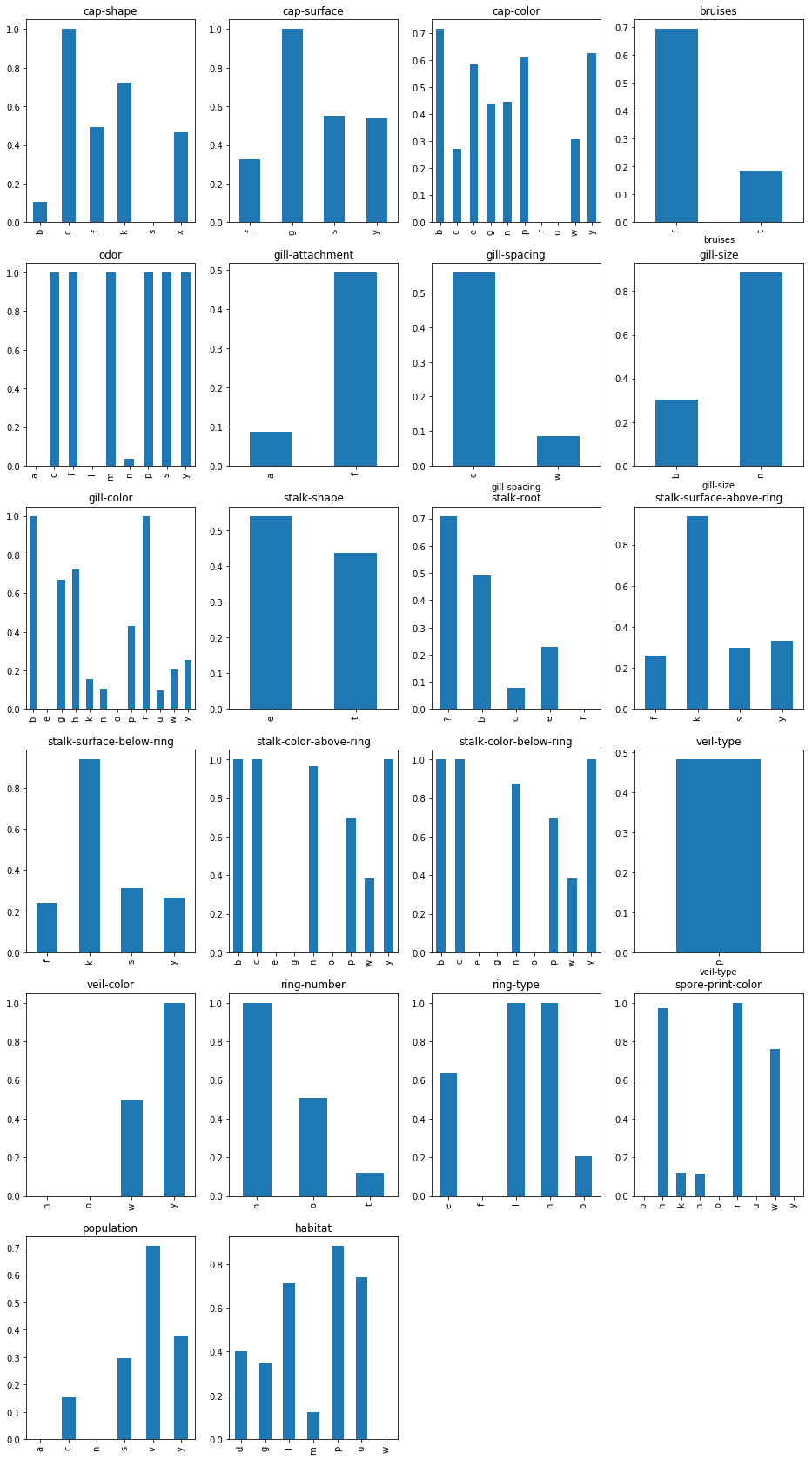
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 plt.close() plt.figure(figsize=(16 ,30 )) i = 1 danger=[] for index_name in dataset.columns[1 :]: result = analysis_poison(dataset[["class" ,index_name]],index_name) ax = plt.subplot(6 ,4 ,i) ax.set_title(index_name) result.plot(kind="bar" ) temp = result[result > 0.75 ] temp = temp.rename(index=lambda x:":" .join([index_name,x])) danger.append(temp) i += 1 plt.show() pd.concat(danger)
c:\users\qiank\appdata\local\programs\python\python35\lib\site-packages\pandas\core\generic.py:3924: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy
self._update_inplace(new_data)
cap-shape:c 1.000000
cap-surface:g 1.000000
odor:c 1.000000
odor:f 1.000000
odor:m 1.000000
odor:p 1.000000
odor:s 1.000000
odor:y 1.000000
gill-size:n 0.885350
gill-color:b 1.000000
gill-color:r 1.000000
stalk-surface-above-ring:k 0.939292
stalk-surface-below-ring:k 0.937500
stalk-color-above-ring:b 1.000000
stalk-color-above-ring:c 1.000000
stalk-color-above-ring:n 0.964286
stalk-color-above-ring:y 1.000000
stalk-color-below-ring:b 1.000000
stalk-color-below-ring:c 1.000000
stalk-color-below-ring:n 0.875000
stalk-color-below-ring:y 1.000000
veil-color:y 1.000000
ring-number:n 1.000000
ring-type:l 1.000000
ring-type:n 1.000000
spore-print-color:h 0.970588
spore-print-color:r 1.000000
spore-print-color:w 0.758794
habitat:p 0.881119
dtype: float64
由上可以发现气味,菌褶颜色,孢子颜色是区分度最大的特征
模型训练——使用决策树模型 数据预处理 特征向量化 1 2 3 model_label = dataset["class" ].replace({"p" :1 ,"e" :0 }) model_dataset = pd.get_dummies(dataset.drop(["class" ],axis=1 )) print(model_dataset.info())
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 8124 entries, 0 to 8123
Columns: 117 entries, cap-shape_b to habitat_w
dtypes: uint8(117)
memory usage: 928.3 KB
None
切分数据集 1 2 3 4 from sklearn.model_selection import train_test_splitx_train,x_test,y_train,y_test = train_test_split(model_dataset,model_label,test_size=0.1 ,random_state=33 ) print(x_train.info()) print(x_test.info())
<class 'pandas.core.frame.DataFrame'>
Int64Index: 7311 entries, 6849 to 7188
Columns: 117 entries, cap-shape_b to habitat_w
dtypes: uint8(117)
memory usage: 892.5 KB
None
<class 'pandas.core.frame.DataFrame'>
Int64Index: 813 entries, 851 to 4472
Columns: 117 entries, cap-shape_b to habitat_w
dtypes: uint8(117)
memory usage: 99.2 KB
None
模型构建 1 2 3 from sklearn.tree import DecisionTreeClassifiertr = DecisionTreeClassifier() tr.fit(x_train,y_train)
DecisionTreeClassifier(class_weight=None, criterion='gini', max_depth=None,
max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, presort=False, random_state=None,
splitter='best')
模型评估 1.0